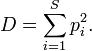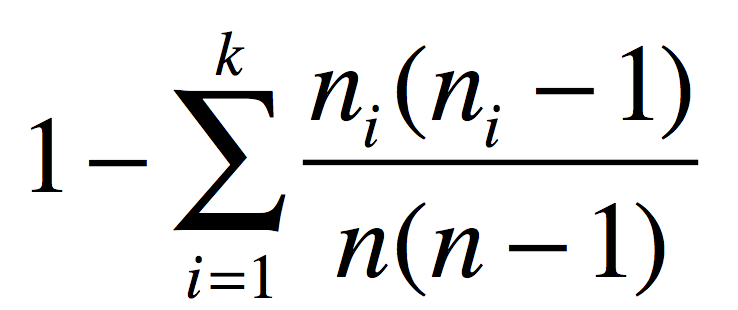PB-SVindex home
PB-SVindex : knowledge-based calculation of Structural Variability Indices of pentapeptide sequences
This tool uses the knowledge of local conformations of pentapeptides in terms of PBs from PB-PENTAdb database to calculate a structural variability index (SVI) for each of them. It shows if a pentapeptide is observed to have multiple local conformations in terms of PBs or not.
This proposed index is similar to the Simpson’s diversity index used in population genetics :
pi is the fraction of all organisms which belong to the i-th species
Here we calculate SVI using an approximation of Simpson’s index :
where, for a given amino acid motif, ni is its occurence in PB class i, and n is the total number of times this motif is found in the database.
This is an estimator for Simpson’s index for sampling without replacement.
Assigned variability index values vary between 0 and 1. Lower values indicate sequences with low structural variability, higher values indicate higher structural variability.
Search by SCOP domain ID:
As output, a table is provided with
- Sequence
- SV index
- Backbone B-factor
- Relative Surface Accessibility (RSA)
Search by entering your amino acid sequences in fasta format (max 1000 sequences) :
As output, a table is provided with
- first 8 characters of ID of sequence (it removes the “>” from first line),
- SV index
with SVI values separated by comas.